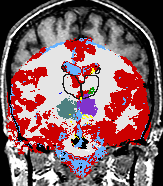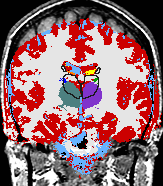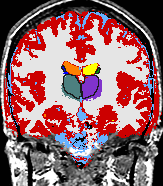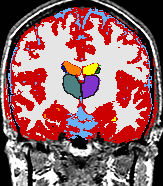
| The Core Mission of the SPL is the extraction of medically relevant information from diagnostic imaging data. |
|
The main SPL laboratory offers drop-in spaces for collaborators. |
The Surgical Planning Laboratory is a computer science oriented laboratory in the Department of Radiology, Brigham and Women's Hospital, which is a teaching affiliate of Harvard Medical School. The SPL does research and development in image processing algorithms, software systems and medical applications. Computer science, digital imaging, and information technologies began finding their way into medicine since the 1950's. However, even today, at the beginning of the 21st century, doctors are not always able to access all of the data about their patients in a comprehensive way. This is particularly true in diagnostic imaging data. For example, it is difficult to assess subtle changes in the volume or shape of a 3 dimensional structure such as a brain tumor or a MS lesion by looking at cross sections only. Today's advanced high-speed imaging devices in some ways only compound the problem of information understanding by hiding potentially life-saving measurements in a torrent of other data. Increasingly, diagnostic imaging scanners are capable to produce not only images of morphology, but are also generating patient specific functional information. While extracting information for clinical purposes may be difficult, the challenge is only multiplied for researchers attempting to learn about diseases such as schizophrenia, multiple sclerosis, or cancer by examining anatomical changes of perhaps thousands of patients over a period of years. Without proper computational tools, medicine cannot fully leverage the potential of imaging for improving the understanding and treatment of diseases that humanity continues to face. |
Short History
| The SPL was founded in 1990 by Ron Kikinis. Ferenc Jolesz was in the process of building the Image Guided Therapy Program at BWH. He had recognized that a successful IGT effort would require sophisticated image post-processing capabilities for integrating pre- and intra-procedure imaging and other information. He recruited Ron Kikinis, who moved from Switzerland to Boston in 1988. Ron Kikinis founded the SPL in 1990. The primary clinical driver for the IGT effort of the SPL was Dr. Peter Black in Neurosurgery. Intraoperative visualization was performed since the founding of the lab. Over the years, a number of other types of interventions were explored, including abdominal, musculoskeletal and cardiac. Several basic technologies were the focus of the initial research activities: Segmentation, Registration, Visualization and Integrated Systems. These technologies can be used not only in IGT but also enable quantitative measures and visualization in a variety of other medical and basic science applications. As a matter of fact, neurologic and psychiatric image analysis were part of the research activities of Ron Kikinis from the first day he joined BWH. High Performance Computing was introduced to the Lab in the early 90's with a IBM PVS system, which was followed by a connection machine from Thinking Machines Corporation. In the late 90's HPC systems from Sun took on this role. The SPL is a research lab without its own degree granting program. This resulted in close cooperation with a variety of schools who sent graduate students into the SPL to do projects in a research oriented clinical environment. In addition, SPL closely collaborated in research with a variety of groups, including academic, and industrial (GE CRD, IBM, Sun Microsystems, MERL). The IGT program added a dedicated OR with an open MR in 1994. In 1997 the SPL won funding for the Neuroimage Analysis Center, NAC. This is a national resource center funded by NCRR. New space in the main campus of BWH was added to enable the expanded effort that this infrastructure grant allowed. |
Early experiments in augmented reality were performed in a collaboration with David Altobelli, Bill Lorensen, Harvey Cline et al.. (US Patent # 5,740,802) |
The SPL's Research
|
|
The main research of the SPL is to develop post-processing methods for digital medical imaging data and to use these methods for answering real-life questions. These scientific and medical applications include, but are not limited to the following examples:
One of the goals of the computational work conducted in the SPL is to develop:
The ability to perform these difficult tasks will allow us to address the computational needs of a wide range of current and future image-based medical applications. While we have still not reached our goal, the current processing time of between a few minutes and a few hours for many tasks allows us to process several hundred studies per year. In addition to a variety of focused grants funding specific research projects there are two center grants that support the SPL's outreach mission and core research. The Neuroimage Analysis Center NAC is a research center supported by the National Center for Research Resources (NCRR) (P41 RR13218) through December, 2011, and presently the National Institute of Biomedical Imaging and Bioengineering (NIBIB) (P41 EB015902) and is focused on the processing of diagnostic imaging data. The National Center for Image Guided Therapy NCIGT is a resource center based on a contractual agreement with NCRR, NCI, and NIBIB and does research into medical procedures enhanced by imaging. The National Alliance for Medical Image Computing NA-MIC is a National Center for Biomedical Computing and part of the NIH Roadmap Initiative and is focused on building an open-source platform for the analysis of medical images. SPL is also participating in two of the BIRN testbeds which focus on multi-site data sharing. |
Examples: The SPL at Work
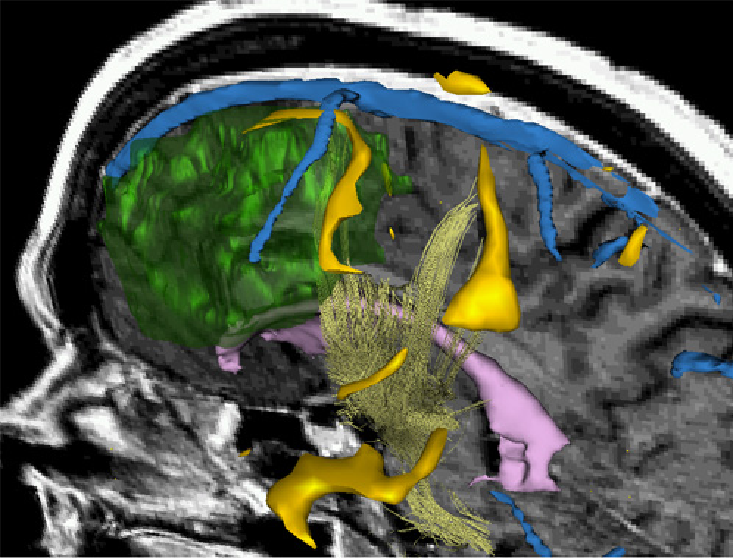
Surgery
|
3D Rendering showing a combination of morphology, fMRI and DTI. This type of visualization can be used to support surgical decisions. Talos et al. MICCAI'03 |
Surgery today relies conceptually on the same principles as it did three thousand years ago: the surgeons use their hands to directly control instruments and they use their eyes to provide them with feedback about the effect of their manipulations. Accordingly, a surgeon needs access to the site of an operation for both reasons: visualization and mechanical access. The modern trend in surgery is an evolution towards minimally invasive approaches, where the damage set for accessing the surgical site is reduced by using rigid or flexible long-necked instruments introduced through natural openings or small incisions into the target areas. These instruments typically carry some form of visualization equipment and some way to introduce instruments for procedures. The big problems arising with this type of approach are:
An additional area of noninvasive treatment methods uses some form of energy deposition in focal hot-spots to prevent access damage at all (e.g. focussed ultrasound). In this scenario, the lack of feedback information for monitoring of the treatment success is even more urgent. Since 1990 we have been providing three-dimensional reconstructions for intraoperative display and monitoring of treatment. By 2006, over 1500 surgeries and procedures have been supported in this way in a variety of fields including neurosurgery, prostate procedures, ENT, liver, other abdominal, cardiovascular. In 1989, the MR division of the Department of Radiology of Brigham and Women's Hospital and Harvard Medical School initiated a project to develop MR-guided interventional procedures. The SPL was founded in 1990 as a spin-off from this effort. In the following years several other components of the image guided therapy program evolved. Today, the National Center for Image Guided Therapies (NCIGT) is the home of these activities. |
Schizophrenia
|
The Schizophrenia Research Project was first discussed in 1987 when Drs. Jolesz, McCarley, and Shenton met to discuss applying MR techniques to investigating brain abnormalities in schizophrenia. The project began in earnest in 1988, when Dr. Shenton received a Career Award (K01) from the National Institute of Mental Health (NIMH) to conduct MRI studies in schizophrenia, and when she and Dr. Ron Kikinis became collaborators. These four investigators have been collaborating on MR studies of schizophrenia ever since, and the project has grown to include a large number of post-doctoral fellows, junior faculty, visiting faculty, and research assistants. These investigators are world leaders in the field of neuroimaging studies of schizophrenia. This project is well funded through grant support from private foundations, NIMH, and the Veterans Affairs Medical Center in Brockton, MA. In 2005, Dr. Shenton moved her primary appointment to Brigham and Women’s Hospital in the Department of Psychiatry, with a secondary appointment in Radiology, and where she founded the Psychiatry Neuroimaging Laboratory. For more information about this project check out the webpages of the PNL. |
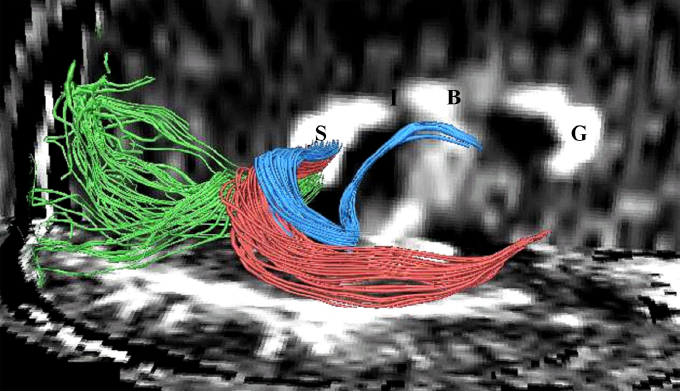
Fibers traveling through the splenium of the corpus callosum are shown, where fibers with different origins have been parcellated using statistical methods. Kubicki et al. N Y Acad Sci. 2005. |
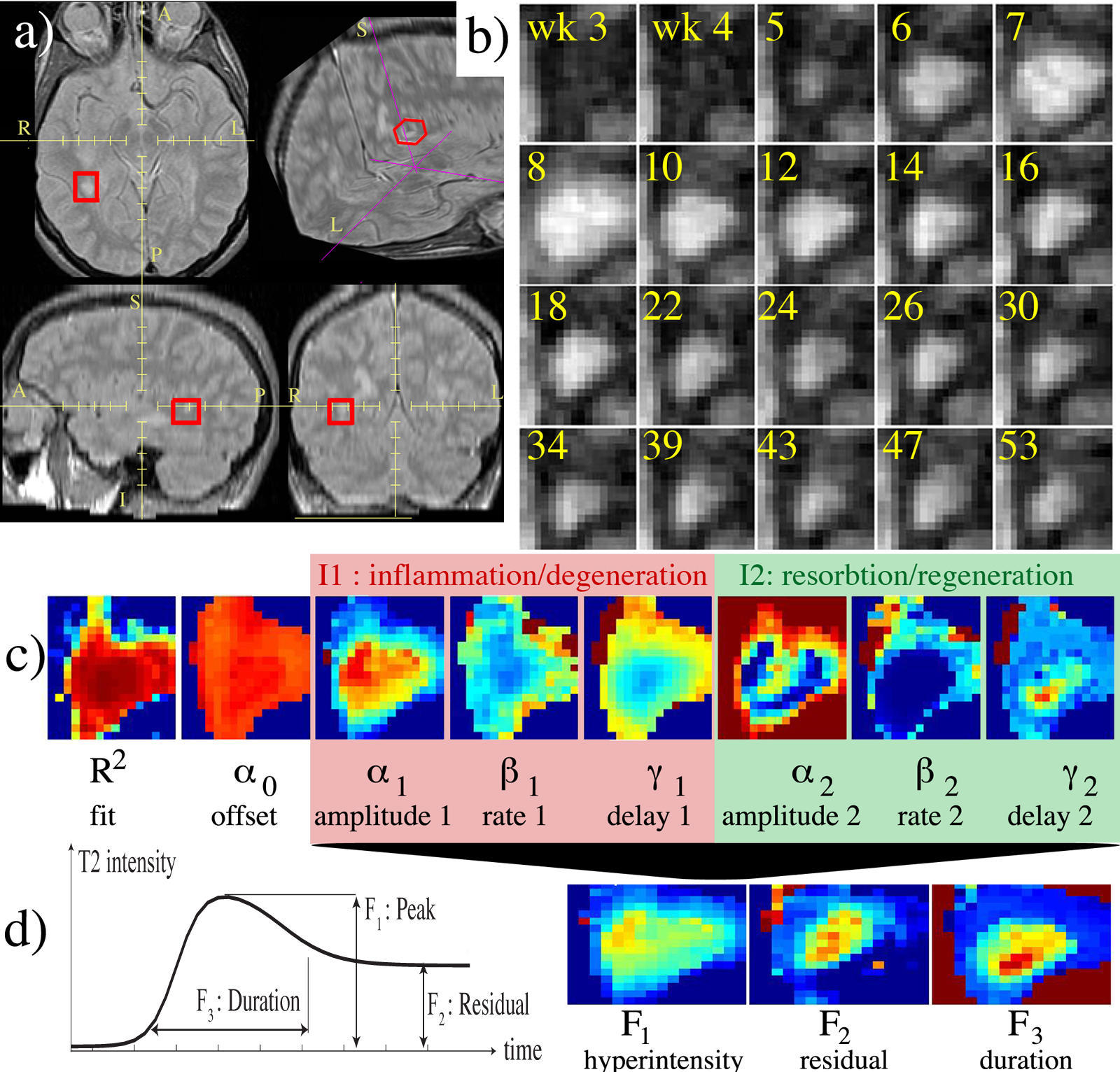
Multiple Sclerosis
|
The temporal evolution of an MS lesion. See Meier et al. Neuroimage 2006 for more info. |
The Multiple Sclerosis (MS) Project at Brigham and Women's Hospital was spun off the SPL in the mid-nineties and became fully operational in early 2000. The CNI is headed by Dr. Charles Guttmann. The purpose of this project is to monitor the progression of MS lesions over a time period of several years. The study required fifty patients to be scanned in the Brigham and Women's Magnetic Resonance Imaging (MRI) scanner twenty four times each. The scans were done over time intervals of from one week to two months in between scans. MS lesions usually appear as bright spots in MRI scans. Our goals were to quantify and categorize these lesions in order to better understand this disease. Our approach to understanding our data was an elaborate one that evolved over time and was initially developed by Dr. Kikinis and Dr. Guttmann, with many novel image processing algorithms supporting the project developed by numerous computer scientists and programmers at MIT Artificial Intelligence Laboratory, and GE Corporate R&D as well as here in the SPL. An image processing pipeline was applied to the datasets generated at each patient visit. The major components of the pipeline are registration and segmentation algorithms. |
Diagnostic Imaging
Currently, MR and CT scanners produce hundreds of megabytes of data per day and unit. Generally, this information is analyzed by a visual evaluation of cross-sectional slices. Increasingly these images are viewed on specialized workstations using sophisticated software packages.
However, it is important to keep in mind that using the above procedures, anatomical structures appear on cross-sectional grey-scale images in a way that is very different from their real appearance. This difference in appearance requires the physician to perform a major mental translation of the information. This translation requires highly specialized training and is very difficult.
While radiologists undergo this specialized training, their clinical partners usually have more problems with the translation. The goal of the work in the SPL is to try to make this job easier by taking the raw imaging data, segmenting out relevant structures, and then generating three-dimensional reconstructions.
This work has several potential applications, such as:
- Making the work of radiologists more efficient by concentrating information from several slices in one rendering.
- Facilitating the communication with referring physicians and to enhance their ability to translate imaging information into a surgical scenario.
- Assisting surgeons in planning for surgical intervention.
- Assisting researchers in the follow up of pathology.
- Assisting in the investigation of small subtle differences in disorders that may not be so apparent from an evaluation of MR or CT scans, such as schizophrenia.
The Future
Diagnostic Imaging
We see an increase of the data production of diagnostic imagers. Just the implementation of methods that are already used in laboratory settings will result in up to two orders of magnitude increase of data production per imager. It can be expected that the unit price of scanners will decrease, while at the same time the reimbursement per scan will decrease as well. This means more data from more scanners and less money to analyze them. This means that the specialists will have to become more efficient in the analysis of diagnostic data and its interpretation by nonradiologists will have to be facilitated because more non-specialists are likely to do some of the reading.
We predict that this will require computerized programs to prescreen the data and point out suspicious areas to the human diagnostician.
Surgery
Minimally invasive procedures will require an increasing amount of visualization in the operating room. While different implementations are thinkable, there is a large common ground.
Preoperatively, a high spatial resolution, high contrast definition diagnostic data set is acquired. The data is segmented and prepared for 3D reconstruction. During the procedure the models are updated using available imaging information. This might be MR images (in the case of the open magnet), plain X-ray images, ultrasound images or video images. The updated models are then used to generate 3D renderings for the surgeon. This can be done using different technologies: monitors in the room, projection into the operation area, projection into a scope (operating room microscope, endoscope), headmounted display.
Computational Challenges
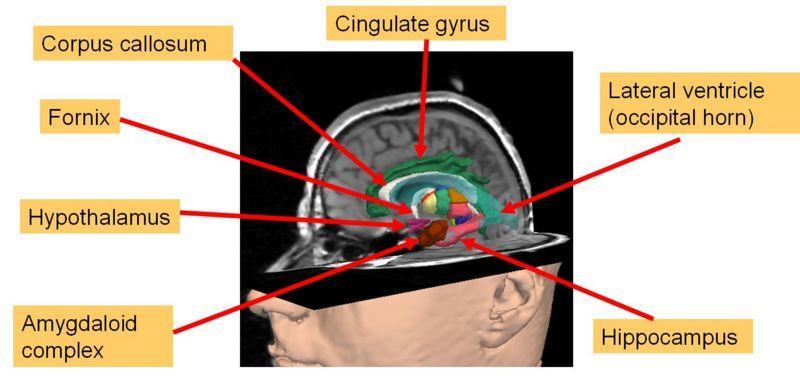
|
Significant work remains to be done both in the algorithmic domain as well as in the computational domain. Some of the classes of segmentation algorithms:
|
Visualization
Our ability to visualize is directly related to our ability to understand; to understand the difficult problems computationally assisted medicine we must have the tools that permit anatomic and abstract data visualization in a way that frees, rather than limits, the physician.
- Multiple streams of real-time stereo rendering for feeding head-mounted displays or other devices for the whole surgical team.
- Real-time update of elastic warps based on interoperative rendering.
- Efficient programs for interactively editing large data sets using haptic input devices.
- These calculations will have to be performed at real-time speeds, which requires optimized hardware and programs.
With the development of new and more powerful computational tools, the SPL will strive to make it possible for doctors to better heal their patients, for scientists to detect and cure disease, and for researchers to better understand the complexity of the human anatomy. In this way, we hope to play a part in the development of the future of medicine.